Alien Life Could Use Endless Array of Building Blocks
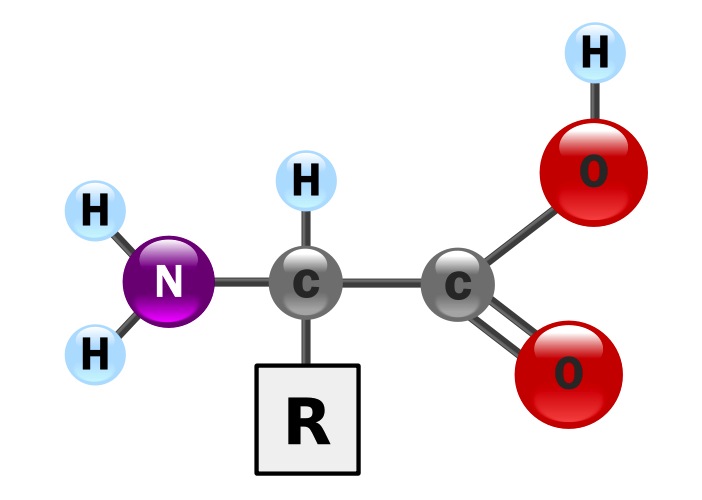
Only 20 standard amino acids are used to build proteins, but why exactly nature "chose" these particular amino acids is still a mystery.
One step toward solving this is to explore the "amino acid space," the set of possible or hypothetical amino acids that might have been used instead. New research has used computer models to construct a large database of plausible amino acids, revealing thousands of amino acid structures that could have been used.
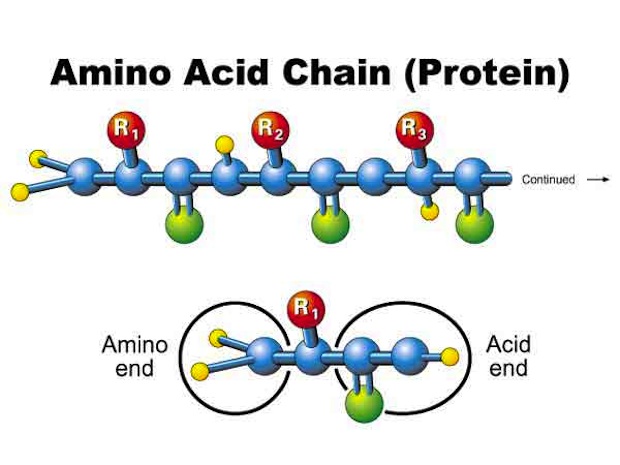
All organisms on Earth employ the same workforce to perform a wide range of essential biochemical tasks. This workforce is comprised of proteins, which are constructed from a long string of amino acids attached to each other. [7 Theories on the Origin of Life]
Even for proteins with particularly long chains of amino acids, there are still only 20 different types of amino acids that are genetically encoded. These amino acids are essentially the building blocks of life, and the same 20 standard amino acids have been used in proteins throughout the history of life on Earth, since the existence of the Last Universal Common Ancestor three to four billions years ago.
Amino acids all have a similar "backbone" structure, which is the foundation upon which the acid is built. This backbone is held together via a single carbon atom acting as a bridge to connect different groups of atoms. Amino acids with a single carbon connector are called alpha amino acids. However, it is possible to have more than one carbon atom in the bridge. In this case, they are called beta amino acids, and so on.
A group of atoms, called a sidechain, is affixed to the backbone, and it is the structure of the sidechain that differs from one amino acid to the next, creating a staggering amount of variability.
Of course, amino acids don't just occur in proteins. There are many more that have different biological functions, and some amino acids are also produced abiotically. Some of these abiotic amino acids are not exclusive to the Earth. For instance, the Murchison meteorite was found to be harboring at least 75 amino acids, and it is even thought that the amino acid glycine might exist in the interstellar medium.
Get the Space.com Newsletter
Breaking space news, the latest updates on rocket launches, skywatching events and more!
However, abiotic chemistry can still only account for half of the 20 genetically encoded amino acids, and there are many unanswered questions as to the role amino acids play. Could extraterrestrial life use a different set of amino acids? Why did life on Earth select those particular amino acids? What other amino acids could have been selected? These are all open questions in astrobiology, and one step toward answering them is to gauge the diversity of the amino acids that could have been used for life on Earth. [What Are the Ingredients of Life?]
Defining amino acids
Markus Meringer, Jim Cleaves and Stephen Freeland set about taking this step by attempting to generate a synthetic map of plausible amino acids structures that are similar in size and composition to the 20 genetic amino acids.
Up until now, modeling these structures has been hampered due to the complexity in generating so many different chemical structures. However, by taking a different approach to the problem, the scientists were able to draw a preliminary amino acid map.
They input a molecular formula into a computer program that had the capability to visualize different amino acid structures based on this formula. However, computing all possible amino acids is a strenuous task for even the fastest computers. Also, listing every possible amino acid does not narrow down the ones of interest to astrobiology. Therefore, the main challenge for the scientists was actually in defining what an amino acid should be, and they used different methods to do this.
Different variations of amino acids
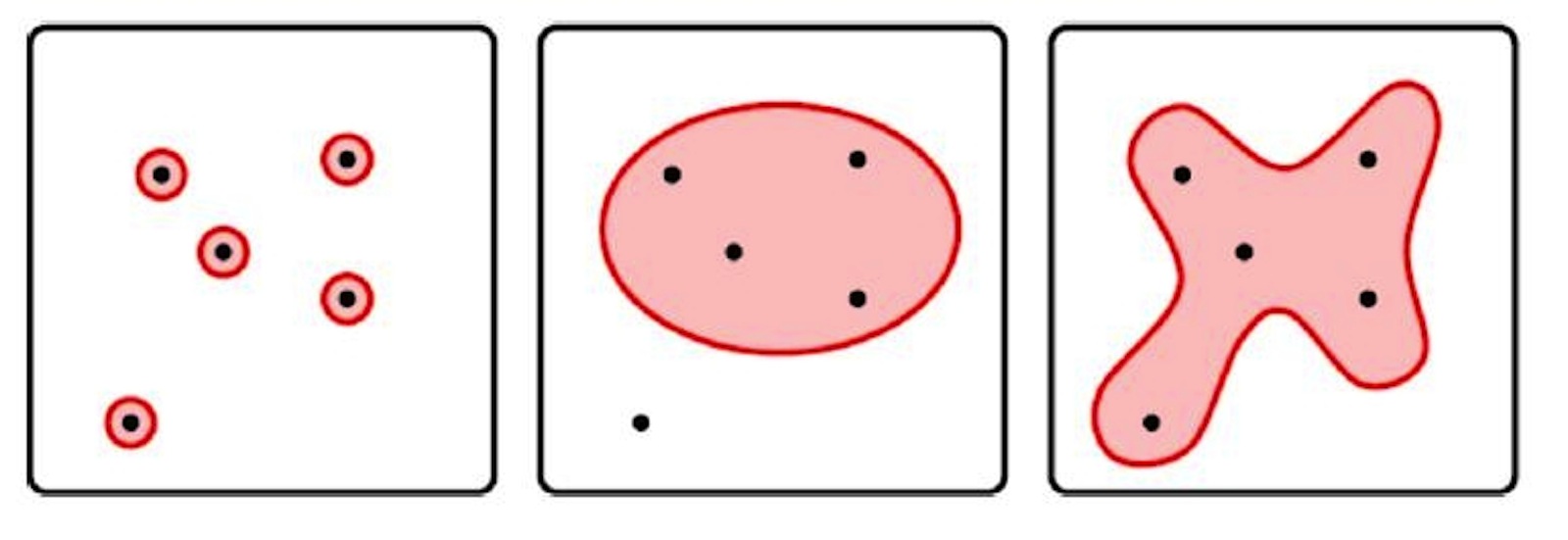
The way to narrow down the interesting amino acids is to explore the "space" around the 20 genetic amino acids. This can be done by generating multiple variations of each amino acid by shuffling the atoms around.
For instance, an isomer has the same molecular formula but a different chemical structure (or arrangement in space), so generating isomers of each amino acid will give the "isomer space."
This isomer space varies in size for each amino acid, partiallydepending on how many atoms there are in the acid. Therefore, the isomer space is largest around tryptophan, the amino acid with the greatest number of atoms.

Fuzzy formulas
However, the isomer space is still a lower limit on the number of potential amino acids that could have been available for use in proteins. The isomer space only probes the area in the immediate vicinity of the amino acid, rather than reaching out toward their neighbors to explore the intervening space between formulas.
Therefore, the scientists included extra combinations by considering the minimum and maximum numbers of possible atoms for each chemical element. The trick that they applied to do this was to use a "fuzzy formula."
This means that instead of telling the software that every atom of every chemical element must occur a certain number of times, the fuzzy formula tells the software to be a bit more vague, or "fuzzy," so that the element can have various numbers of atoms. For example, oxygen could be specified as a range from 2 to 4, so that the program would search for solutions that included 2, 3 or 4 oxygen atoms.
Using this fuzzy formula uncovered a treasure trove of additional amino acid combinations. However, a single fuzzy formula can only be used to explore the space around 15 of the amino acids. A single formula that can include all 20 is still too much for current computing power to handle.
Biochemistry's palette
The next step was to try and explore the amino acid space beyond the isomers while including the five that had been neglected in the previous step. This meant that multiple fuzzy formulas had to be used, but this couldn't be done without classifying the genetic amino acids into 10 different groups.
"There a lot of ways one could classify the coded amino acids according to functional groups and properties," Cleaves said. "But if you stuck to just using the functional groups observed in biology and computationally poked around with that chemical diversity, it wouldn't be nearly as wide as what we came up with, and it's clear that biochemistry had a huge palette to play with during evolution."
Using 10 fuzzy formulas proved to be the most successful way of exploring the amino acid space. Not only does this method have less processing time than using one fuzzy formula, but it has the advantage of including variations of all of the genetic amino acids. [Genetics by the Numbers: 10 Tantalizing Tales]
Cartography of amino acids
The number of amino acid structures generated surpasses all previous estimates. Using the method with the single fuzzy formula produced 120,000 plausible structures, and using 10 fuzzy formulas narrows this down to a more biologically relevant set of nearly 4,000 amino acids. This shows that there were a staggering number of options available that could have possibly been used for building the genetically encoded amino acid set — and yet there are only 20.
They compared the output of both methods to databases of biological alpha amino acids beyond the 20 genetic ones, as well as to amino acids found in carbonaceous meteorites. Many of the amino acids present in the computer library also occur in nature, showing that the computer generation of amino acids is a way of identifying potentially interesting amino acids that could be used in proteins. It is even possible that there are undiscovered natural amino acids that have had their chemical structure probed by the computer database.
The computer libraries generated by the team can now be used as a foundation for further exploration into the jungle of amino acids, and may ultimately lead to an understanding of life's building blocks.
The research was published in the November issue of the Journal of Chemical Information and Modelingand can be found here: http://pubs.acs.org/doi/abs/10.1021/ci400209n
This story was provided by Astrobiology Magazine, a web-based publication sponsored by the NASA astrobiology program.
Follow us @Spacedotcom, Facebook or Google+. Published on SPACE.com.
Join our Space Forums to keep talking space on the latest missions, night sky and more! And if you have a news tip, correction or comment, let us know at: community@space.com.
Amanda Doyle is a freelance science writer who studied astrophysics at Keele University in the United Kingdom, earning a Ph.D. for her studies into the spectral analysis of solar-like stars. Her work as appeared in All About Space Magazine, BBC Sky At Night and Astronomy Now. She is a former editor of Popular Astronomy and has served as Deputy Transport Editor at Chemical Watch news & events by Enhesa.










